AIR
-
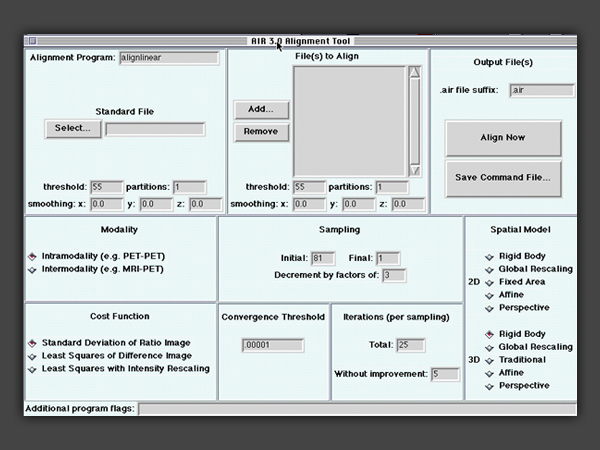
AIR

Automated Image Registration
Description
AIR allows automated registration of 3D and 2D images within and across subjects and within and sometimes across imaging modalities. AIR source code is written in C. The software also includes a graphical user interface, written in Tcl/Tk, for two of the most frequently used programs. The AIR package is distributed only as source code (no executables). In addition, a number of utility routines and error checking routines are provided. The AIR library can be used to easily incorporate automated image registration into site-specific programs adapted to your particular needs.
Visit the AIR website.
Features
• The alignlinear program includes 2D and 3D variants of all linear spatial transformation models and supports within-modality, across-modality and linear intersubject registration.
• The alignment program, align_warp, includes 2D and 3D variants of nonlinear polynomial spatial transformation models ranging from first (linear) to twelfth order.
• The alignment program, align_warp, includes 2D and 3D variants of nonlinear polynomial spatial transformation models ranging from first (linear) to twelfth order.
• The program manualreslice can be used to specify values for roll, pitch, yaw, x_shift, y_shift, and z_shift, x-axis_scaling, y-axis_scaling and z-axis_scaling.
Purpose
The AIR package includes automated routines for aligning and reslicing tomographic image data, including intrasubject, intramodality registration (e.g., PET to PET or MRI to MRI); intrasubject, intermodality registration (e.g., PET to MRI); and intersubject registration (e.g. subject to subject or subject to atlas template) Intrasubject registration of brain images uses a rigid-body model. Intermodality registration has been validated for some (MRI-PET), but not all modalities. Intersubject registration can be performed using any of a variety of linear or nonlinear models to register different subjects to one another or to an atlas template (for example, an averaged brain in "Talairach space"). These models may also be useful for intrasubject registration of organs that are more deformable than brain or for tracking intrasubject developmental changes over time. In addition to 3D models, AIR includes homologous 2D deformation models that may be useful in selected circumstances (e.g., single slice fMRI data, pure 2D images, etc.).
Installation
UNIX (including Linux and Mac OS X)
UNIX configuration and compilation instructions:
• In the main AIR directory, type './configure'. This will execute a shell script that should automatically create three files. The first of these will be in the main AIR directory and is called 'Makefile'. The other two will be in the src/ subdirectory and are called 'Makefile' and 'config.h'.
• If ./configure fails, it should print out some insight into the problem that you may be able to fix. Alternatively, you might consider some of the workarounds listed below
• If you troubleshoot ./configure, you sometimes need to remove ./configure.cache to get ./configure to forget results of earlier runs
• Caution To Developers: the Makefiles that are generated do not contain the level of dependency information needed for efficient software development. Developers should consider building a shared library from the src directory subroutines and linking to this shared library as a strategy for shifting such dependencies to runtime.
• In the src/ subdirectory, edit the 'config.h' file to meet any site specific preferences.
• Any text editor can be used (be sure to save any changes as plain text), configuration details are provided below.
• Optionally, review and edit the two files called 'Makefile'
• In the main AIR directory, type 'make all'. This should compile the code
• If you want the compiled programs moved to the bin subdirectory, type 'make install' in the main AIR directory
• If you ultimately want the programs somewhere else entirely, it is much easier to copy them from /bin, so type 'make install' to move them to /bin first
• Before rebuilding AIR (e.g., after changing something in src/config.h) you must type 'make clean' in the main AIR directory before typing 'make all' again 'make clean' removes all of the compiled libraries and programs so that new, internally consistent versions can be built
• To start over from the beginning at any point, type 'make distclean' in the main AIR directory and return to step 1
• 'make distclean' removes all of the compiled libraries and programs as well as the configuration information and Makefiles
PC (DOS, Windows and Windows NT)
Please note that different PC compilers have different requirements and only general recommendations are provided here.
• In the src subdirectory, copy the file 'config_PC.h' to 'config.h'
• Edit the 'config.h' file to meet any site specific preferences
• Any plain text editor can be used.
• In the main AIR directory, edit the files AIRmain.h and HEADERmain.h, removing the lines with UNIX style forward slashes and uncommenting the lines with the PC style backslashes
• After modification, AIRmain.h should read: #include "src\AIR.h"
• After modification, HEADERmain.h should read: #include "src\HEADER.h"
• There should not be any spaces or tabs before '#include' in either of these files
• Using your PC C compiler, compile all of the files in the src subdirctory into a library. Consult your C compiler documentation for details
• Using your PC C compiler, compile each of the programs in the main AIR directory. For example, compile the file 'alignlinear.c' to generate the executable file 'alignlinear.exe'. You will need to link in the library compiled in the previous step.
• Consult your C compiler documentation for details
• Note that the files src/AIR.h, src/config.h and src/HEADER.h are required when compiling each of the programs in the main directory.
Grants & Acknowledgments
NIH-NCRR 9P41EB015922-15 and 2-P41-RR-013642-15
NIH-NINDS P01-NS15654
NIH-NINDS 1 K08 NS01646
NIH-NIMH R01-MH37916
DOE DE-FC03-87ER 60615


